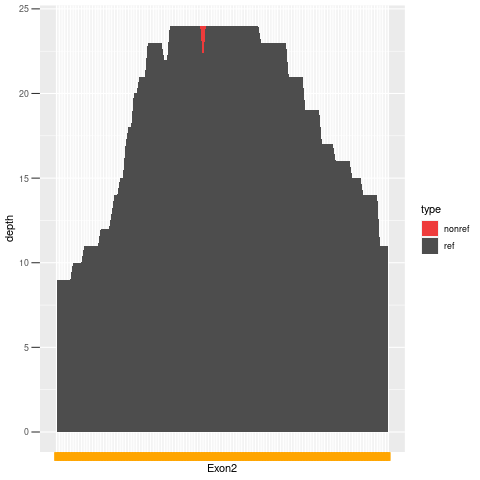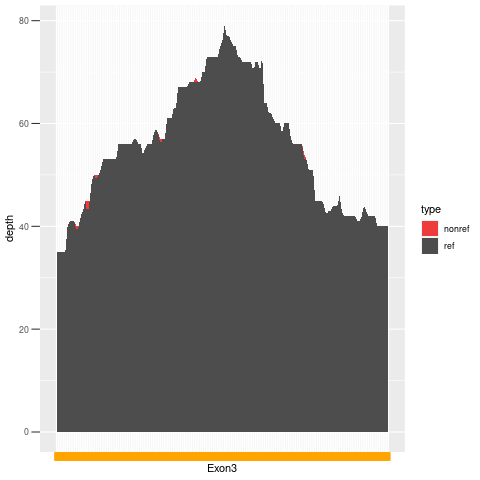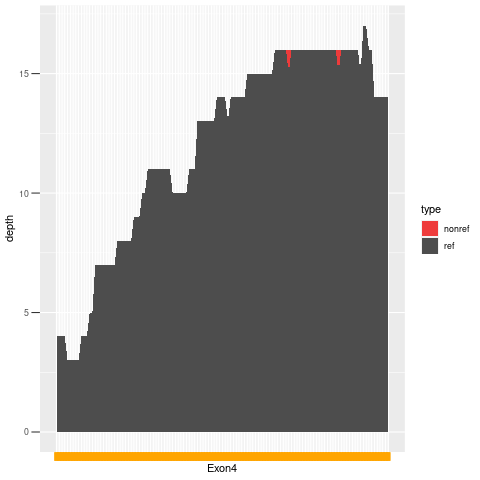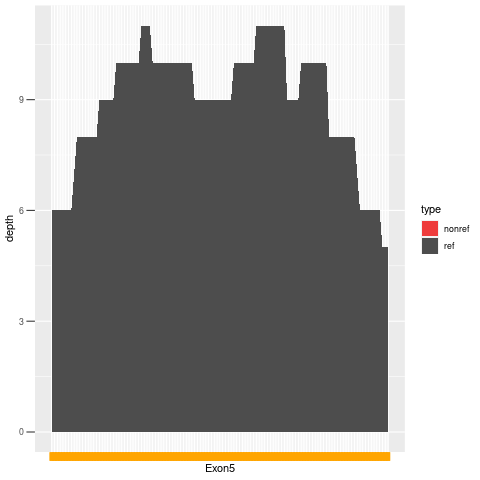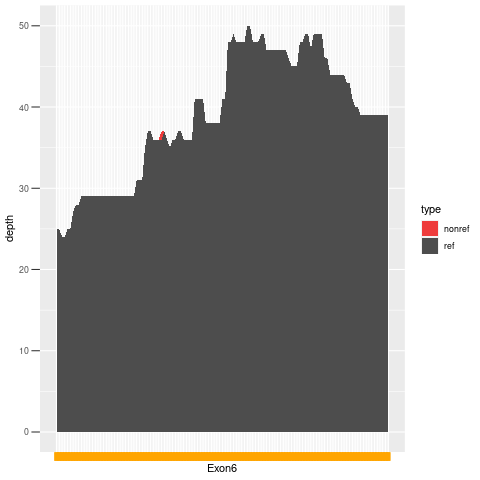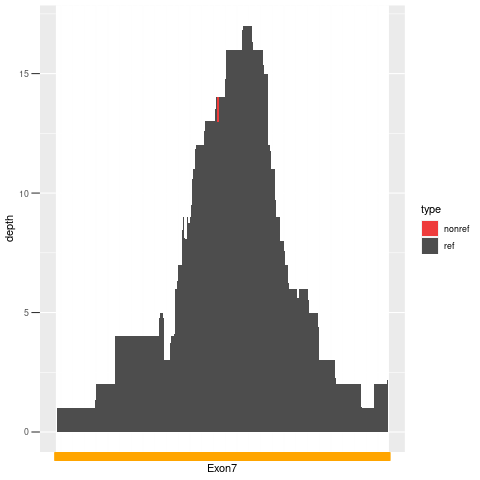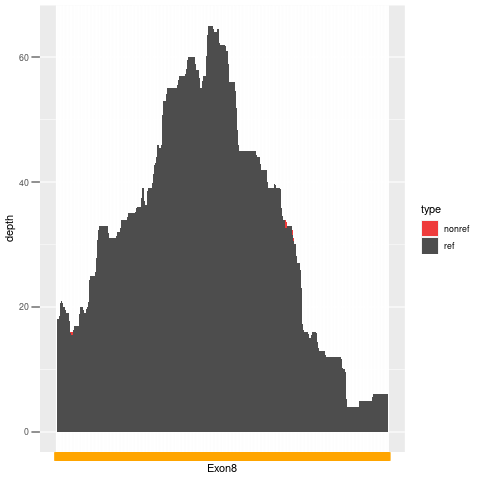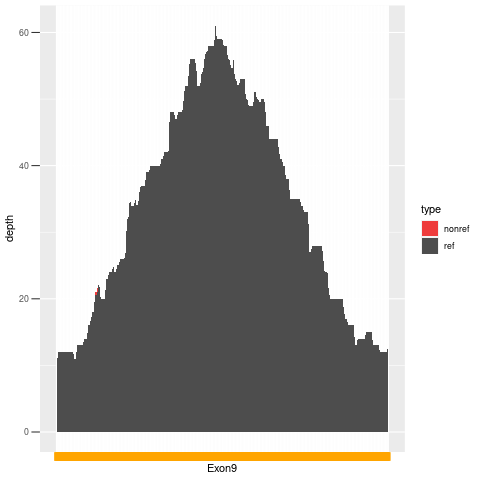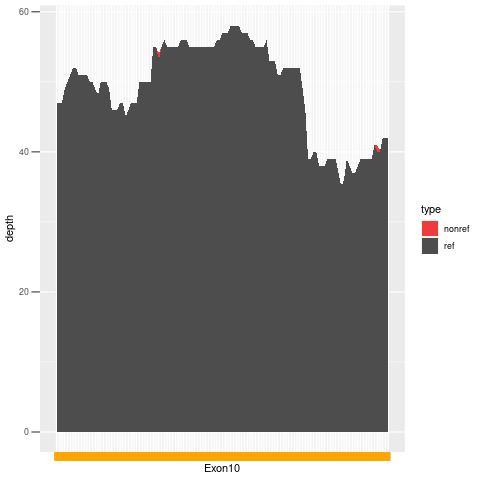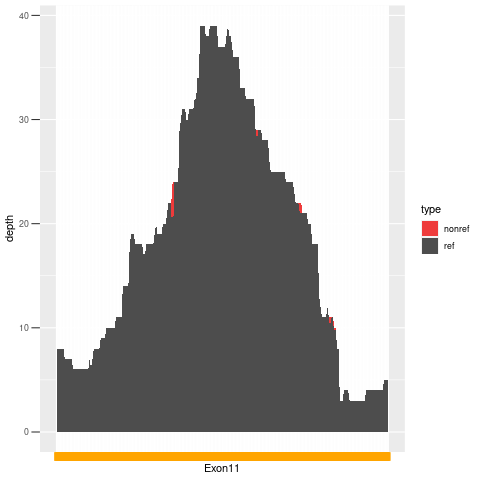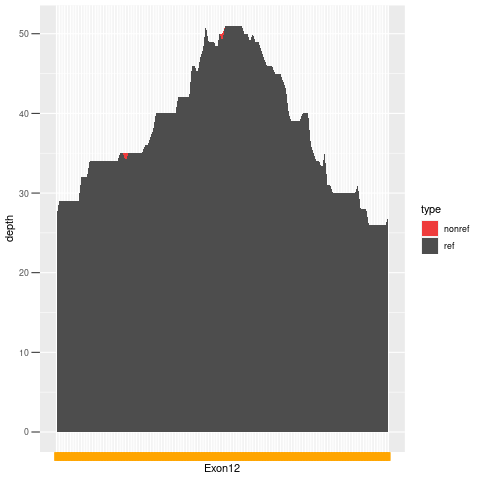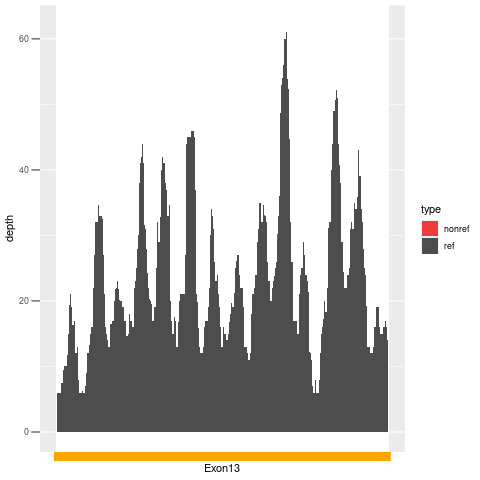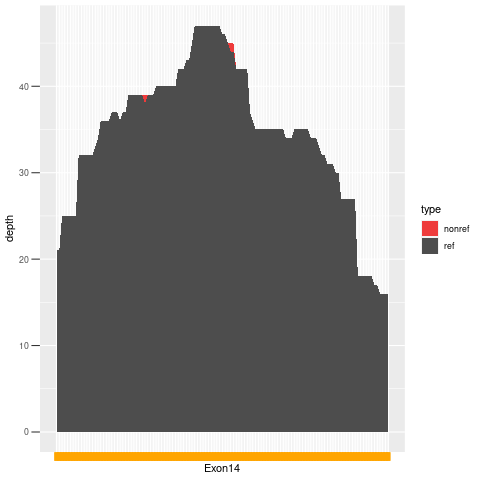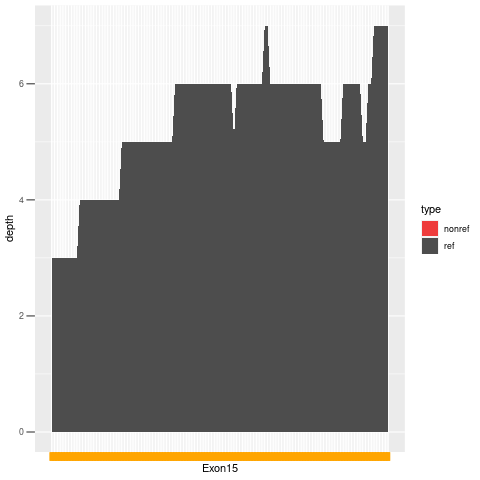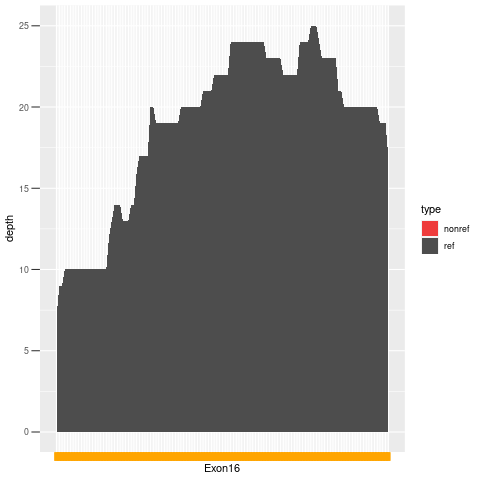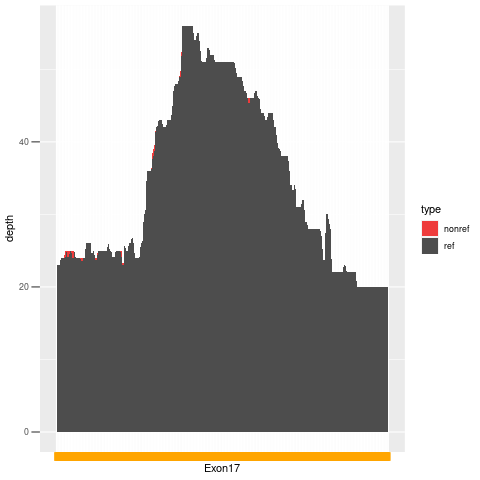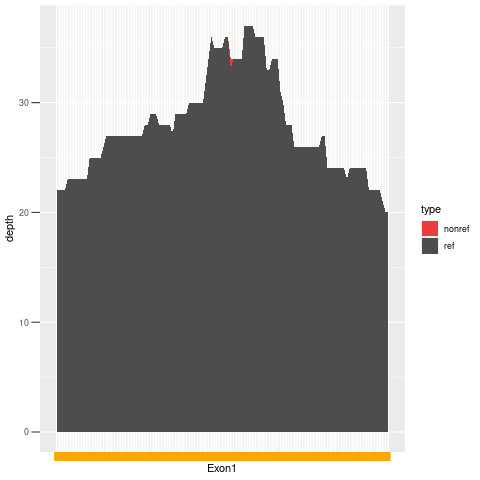
| Exon no | Exon1 |
| Genomic coordinates | chr17:41201114-41201234 |
| Exon length (bp) | 121 |
| GC% | 44.63 |
| Mean depth of coverage | 27.83X |
| % covered with at least 1X depth | 100.00 |
| % covered with at least 5X depth | 100.00 |
| % covered with at least 10X depth | 100.00 |
| % covered with at least 20X depth | 100.00 |
| % covered with at least 30X depth | 28.93 |