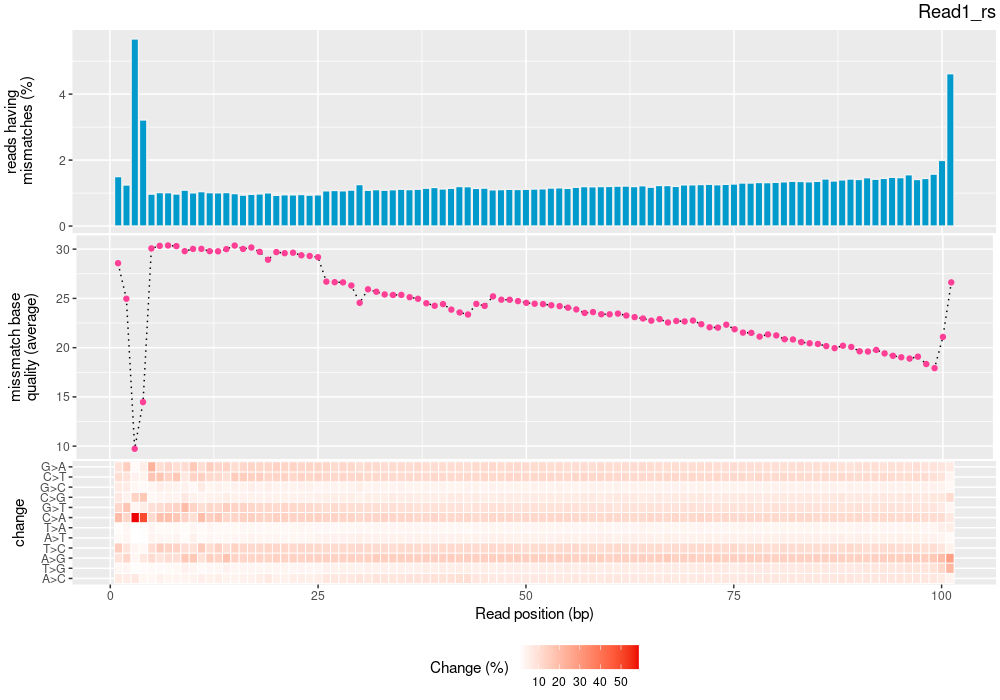
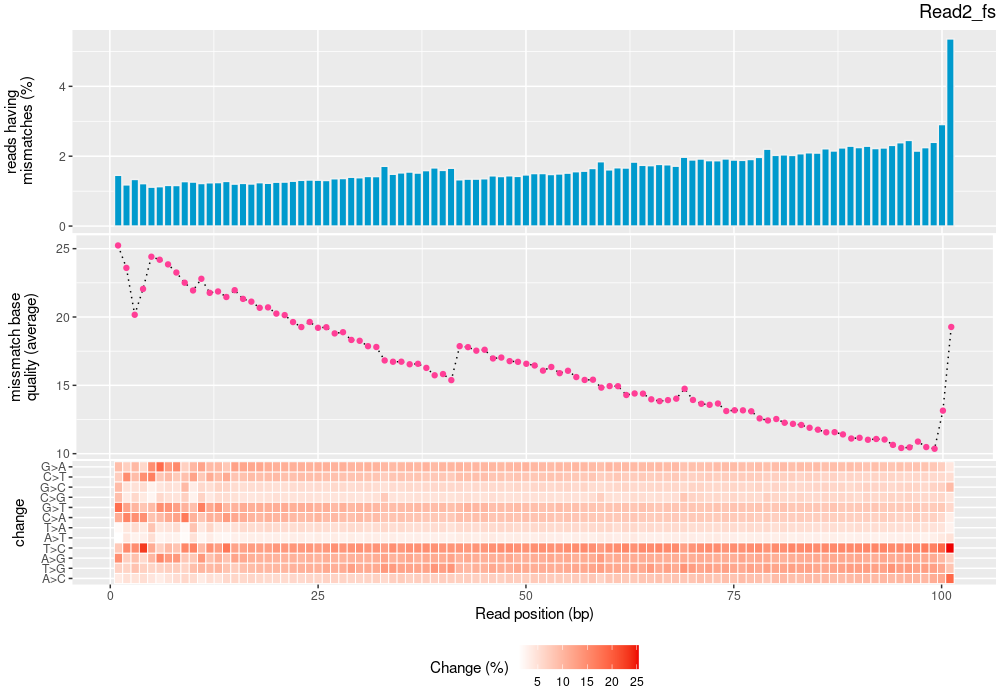
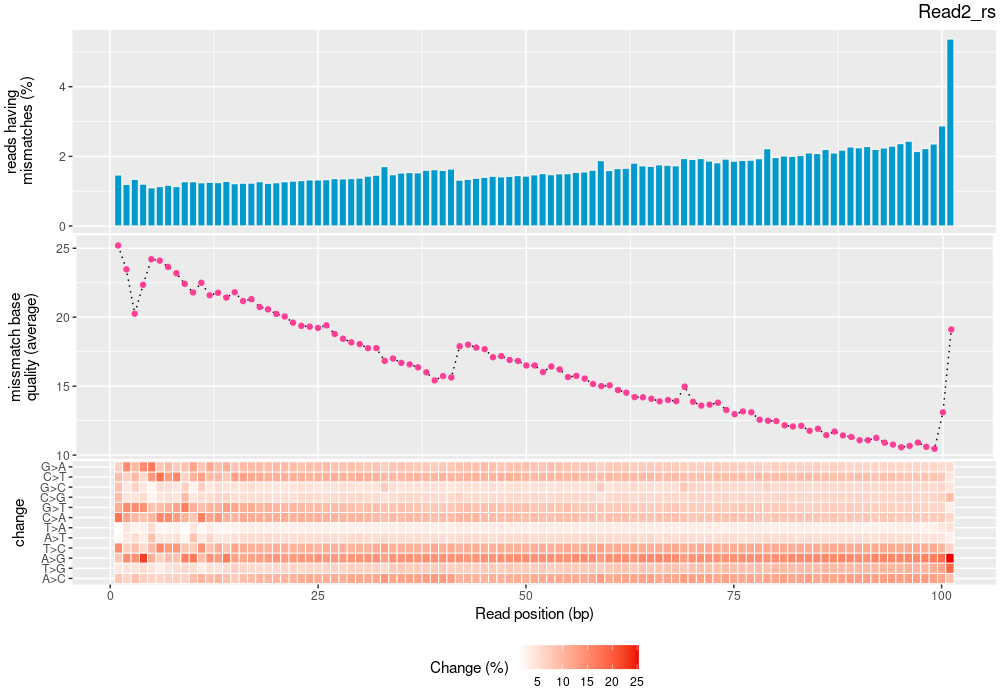
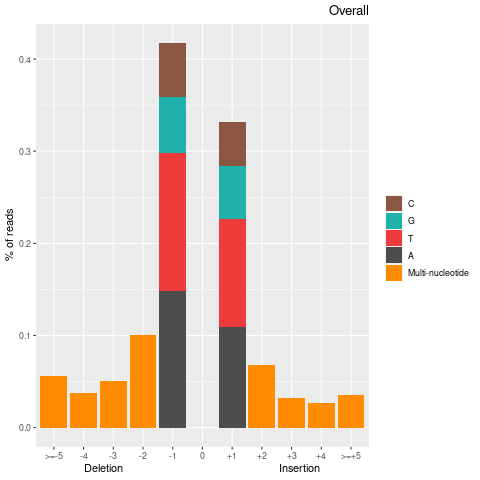
| mapinsights bamqc -r reference.fa -i input.bam -o bamqc-output/ |
| Alignment file | input.bam |
| Reference file | reference.fa |
| Bed file | Not specified |
| No of contigs in alignment file | 86 |
| No of @RG tag | 4 |
| Sample name [@RG SM] | NA12878_CGTACTAG_L1 |
| Library [@RG LB] | na12878 |
| Identifier [@RG ID] | NA12878_CGTACTAG_L1 NA12878_CGTACTAG_L2 NA12878_TAAGGCGA_L1 NA12878_TAAGGCGA_L2 |
| Analysis date | Fri Apr 8 15:38:39 2022 |
| Homopolymer size | 5 |
| Reference size (bp) | 3137454505 |
| Targeted region size (bp) | 0 |
| No of reads in input file | 34321158 |
| Unmapped reads | 7706 (0.02%) |
| Overall | Read1 | Read2 | |
| Mapped-reads | 34313452 | 17067200 (49.74%) | 17246252 (50.26%) |
| Mapped-read1 | 17067200 (49.74%) | 17067200 (100.00%) | - |
| Mapped-read2 | 17246252 (50.26%) | - | 17246252 (100.00%) |
| Mapped-forward | 17159908 (50.01%) | 8536942 (49.75%) | 8622966 (50.25%) |
| Mapped-reverse | 17153544 (49.99%) | 8530258 (49.73%) | 8623286 (50.27%) |
| Mapped-pair | 34313452 (100.00%) | 17067200 (49.74%) | 17246252 (50.26%) |
| Mapped-properpair | 33725003 (98.29%) | 16821228 (49.88%) | 16903775 (50.12%) |
| Secondary-alignments | 627972 (1.83%) | 220624 (35.13%) | 407348 (64.87%) |
| Supplementary-alignments | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| QC-failed | 0 (0.00%) | 0 (0.00%) | 0 (0.00%) |
| Strand-ratio (F:R) | 0.50:0.50 | 0.50:0.50 | 0.50:0.50 |
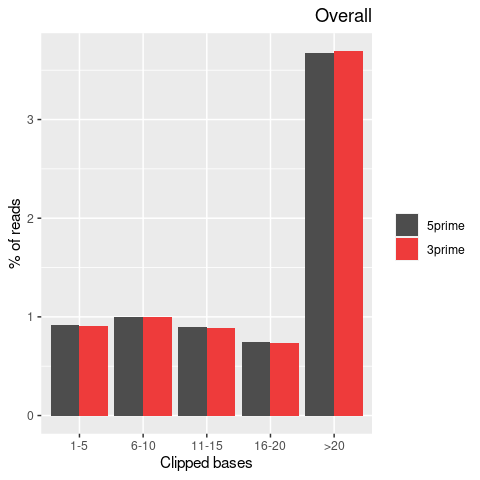
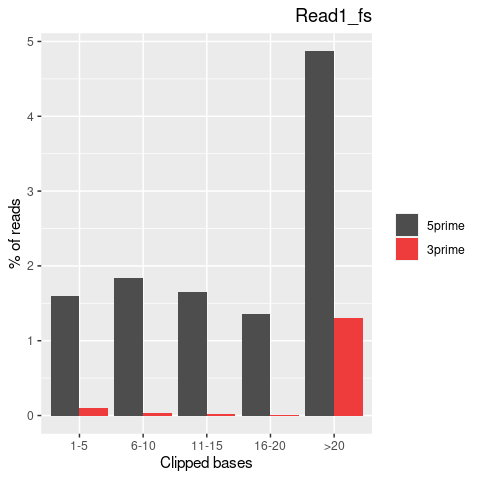
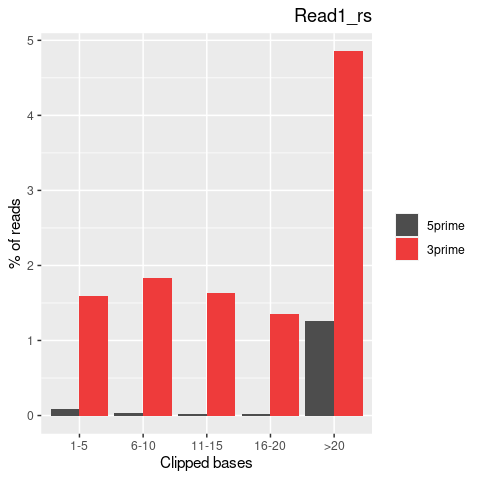
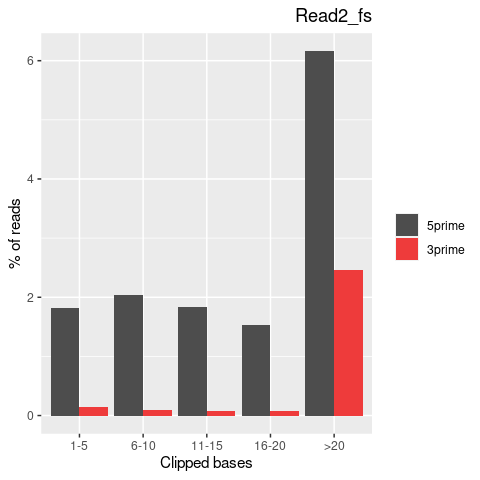
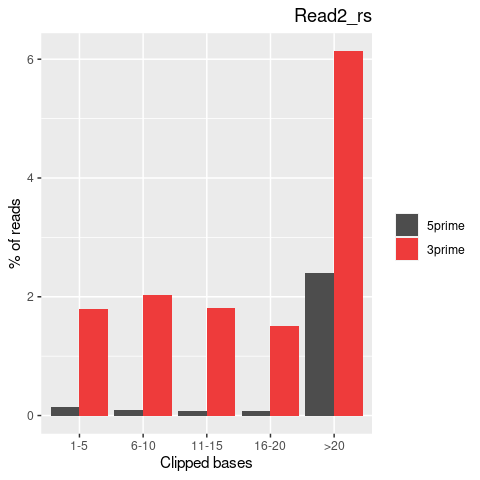
| Softclip-events | 3808025 | 1767710 (46.42%) | 2040315 (53.58%) |
| Softclipped basecounts | 81837644 | 36528879 (44.64%) | 45308765 (55.36%) |
| Hardclip-events | 1150583 | 405725 (35.26%) | 744858 (64.74%) |
| Hardclipped basecounts | 50404797 | 17765769 (35.25%) | 32639028 (64.75%) |
| Illumina-Adapter | 1019 | 857 (84.10%) | 162 (15.90%) |
| Illumina-PCR-primer | 850 | 849 (99.88%) | 1 (0.12%) |
| Nextera-Transposase-Sequence | 851835 | 427176 (50.15%) | 424659 (49.85%) |
| A | 839117306 (25.18%) | 420967792 (50.17%) | 418149514 (49.83%) |
| T | 841379466 (25.24%) | 422076928 (50.16%) | 419302538 (49.84%) |
| G | 826352077 (24.79%) | 413172072 (50.00%) | 413180005 (50.00%) |
| C | 825969914 (24.78%) | 413070230 (50.01%) | 412899684 (49.99%) |
| N | 269354 (0.01%) | 43771 (16.25%) | 225583 (83.75%) |
| GC % | 49.58 | 49.50 | 49.66 |
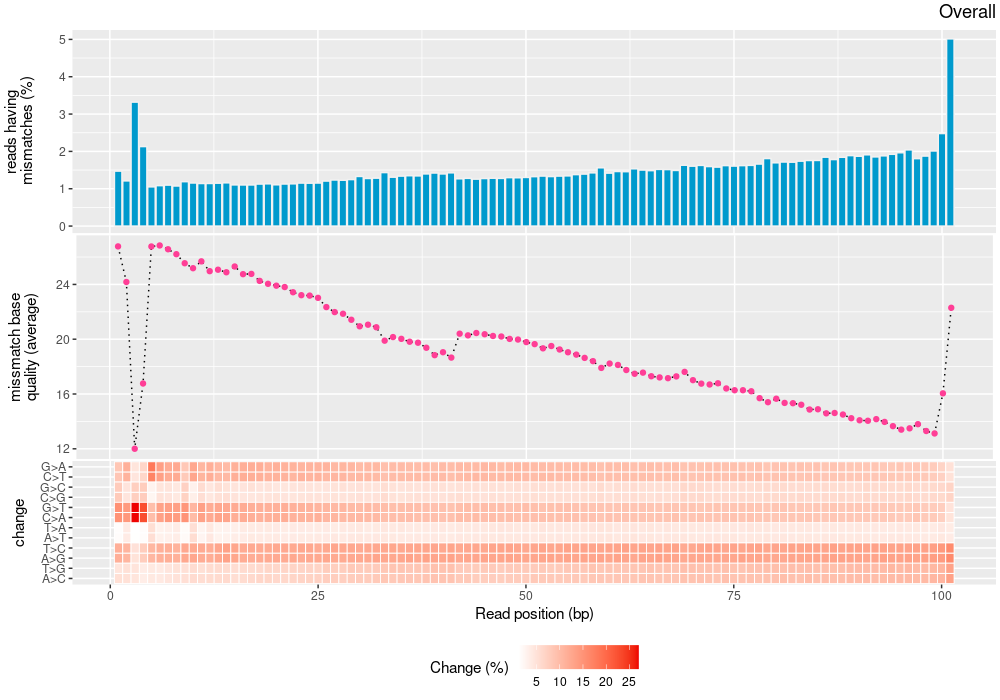
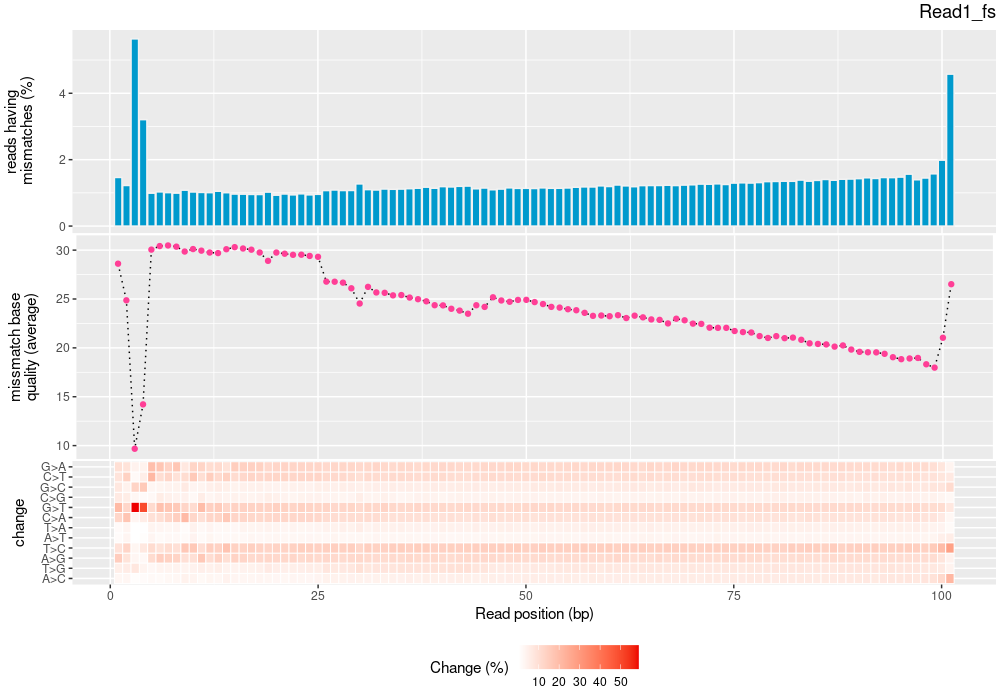
| Single nucleotide match | 3323886859 | 1665920315 | 1657966544 |
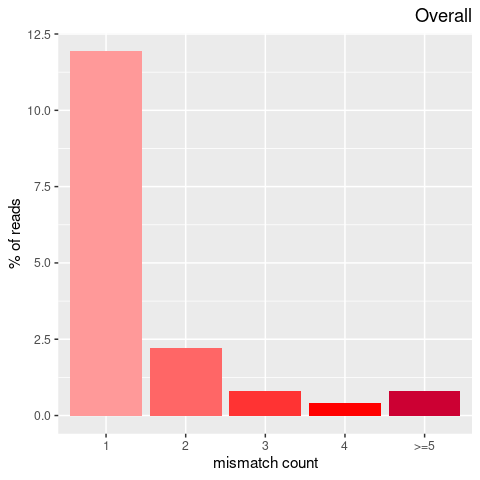
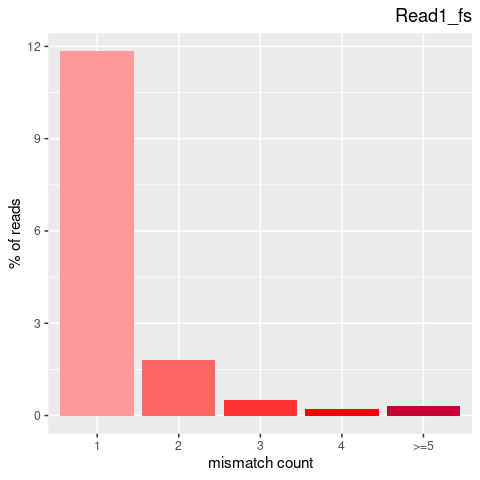
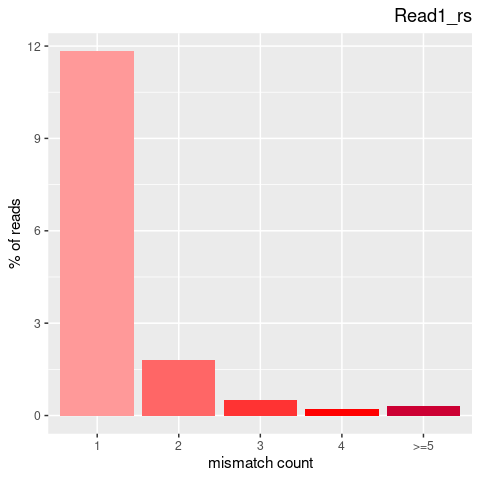
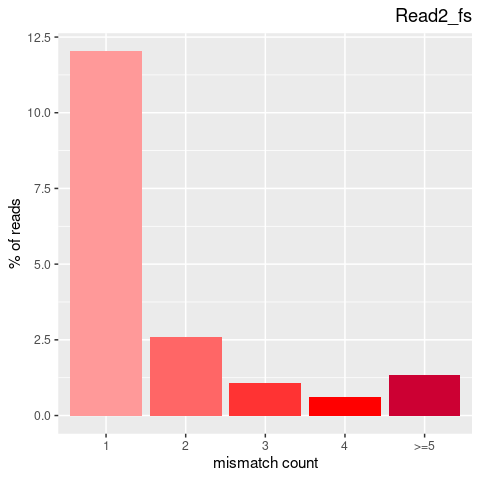
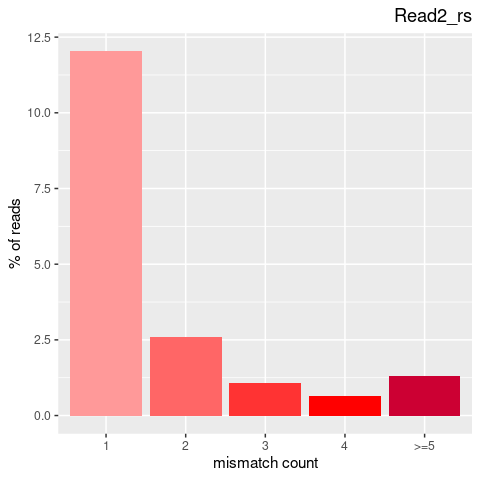
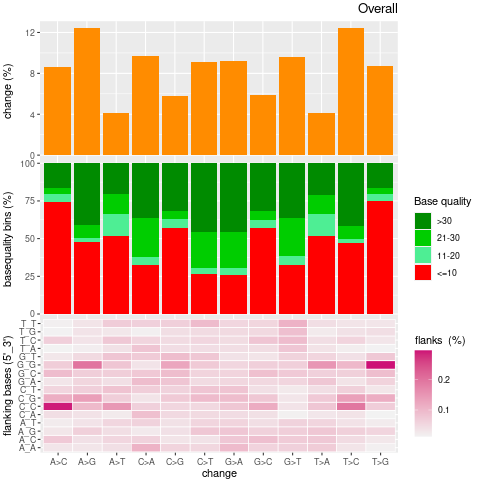
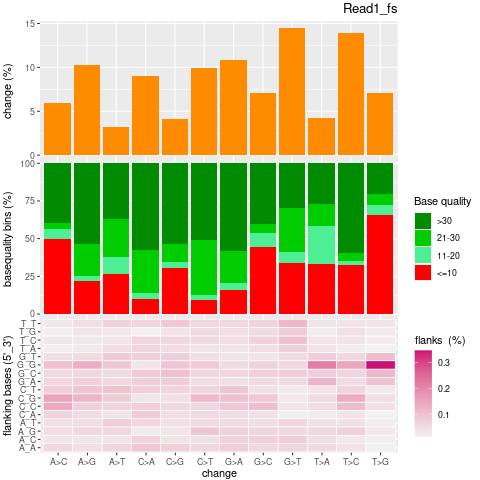
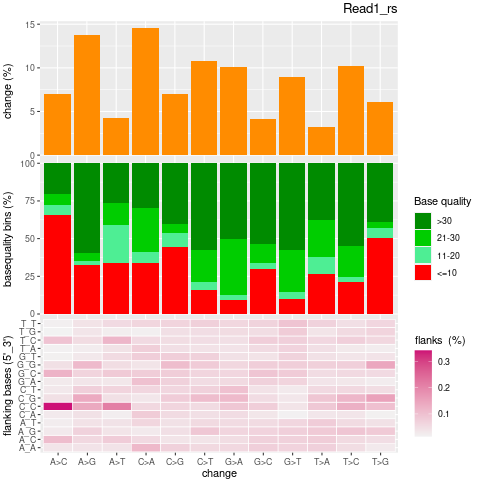
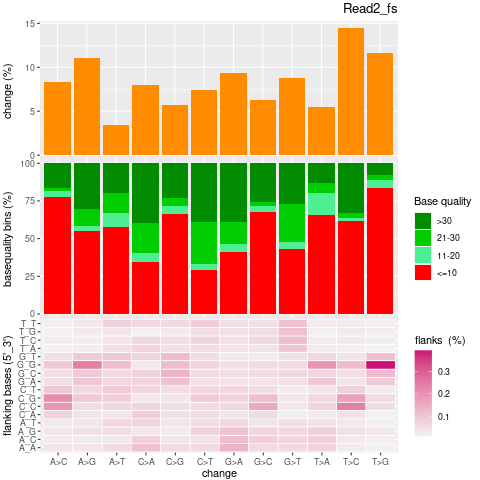
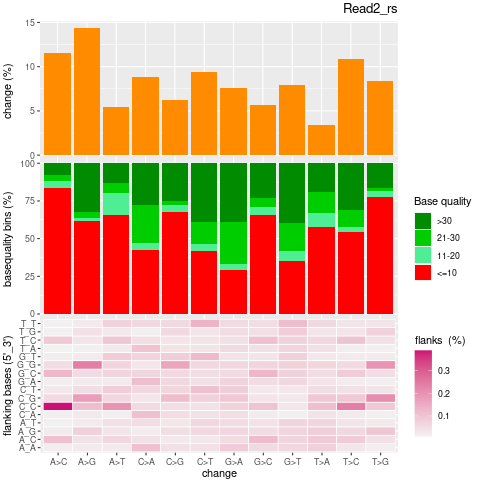
| Single nucleotide mismatch | 8931904 | 3366707 | 5565197 |
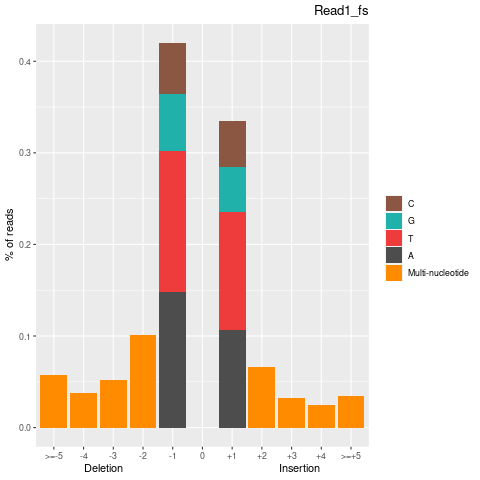
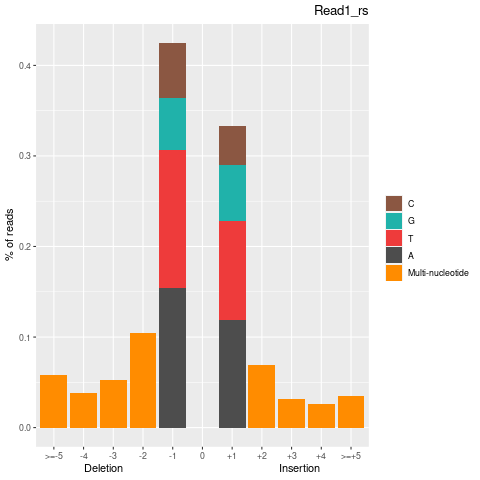
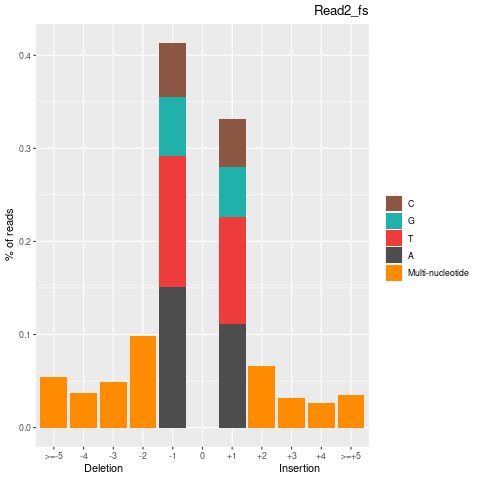
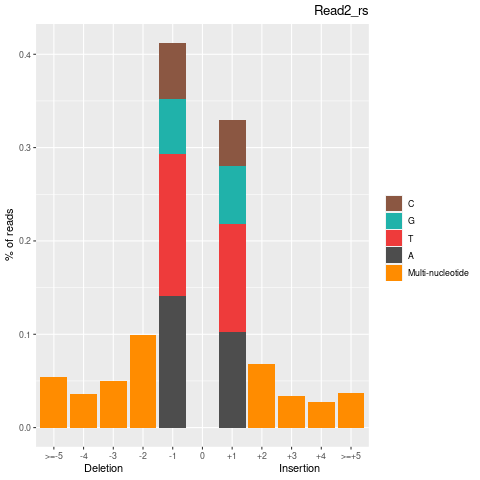
| Insertion-events | 170177 | 84635 | 85542 |
| Inserted basecount | 328094 | 161759 | 166335 |
| Homopolymer Insertation | 77398 | 39491 | 37907 |
| Deletion-events | 227957 | 115151 | 112806 |
| Deleted basecount | 494580 | 250382 | 244198 |
| Homopolymer Deletion | 90326 | 46356 | 43970 |
| Mismatch rate | 0.0027 | 0.0020 | 0.0033 |
| Insertion rate | 0.0001 | 0.0001 | 0.0001 |
| Deletion rate | 0.0001 | 0.0001 | 0.0001 |
| Mapped-diffChr | 737807 (2.15%) |
| Insertsize>=1K | 138272 (0.40%) |
| Mapped-pair forward | 221269 (0.64%) |
| Mapped-pair reverse | 222373 (0.65%) |
| Mapped-pair outward | 46522 (0.14%) |
| Mapped-pair innerward | 33823288 (98.57%) |
| Mean depth of coverages | 1.06X |
| DuplicateMarked rate | 2148701 (6.26%) |
| Mean insertsize | 182.0 |
| Mean base quality | 31.9 |
| Mean mapping quality | 57.9 |
| Mean read length | 99.5 |